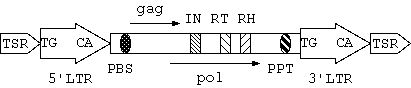
Figure 1: basic structure of a full-length LTR retrotransposon
The typical structure of a full-length LTR retrotransposon is shown in Figure 1:
~6bp short
direct repeat string flanking the 5' and 3' extremities of an
element. It is the sign of insertion of transposable elements.
~18bp
sequence complemented to the 3' tail of some tRNA. The site is very
important because tRNA binding process is the first step of
initiating reverse transcription.
~15 bp in length. Like PBS, this region is important
for reverse transcription.